The Leguminosae is one of the most biodiverse families of angiosperms, with nearly 20,000 species, including many agriculturally and economically important crops. Leguminosae can be divided into six subfamilies, among which Bauhinia subfamily (Cercidoideae) has 12 genera and more than 300 species, and Glycyrrhizae subfamily (Detarioideae) has 84 genera and more than 700 species, which is the earliest branch of Leguminosae that was differentiated 67 million years ago; however, the lack of genomic information on these two subfamilies has greatly limited systematic genomic studies of leguminous plants, in particular, inferring the origin of new genes. especially the inference of the origin of new genes.
Recently, Qi Ji’s team from Fudan University published a paper entitled “The nearly complete assembly of Cercis chinensis genome and Fabaceae phylogenomic studies” in Plant Communications. The nearly complete assembly of Cercis chinensis genome and Fabaceae phylogenomic studies provide insights into new gene evolution”, which greatly facilitates the systematic genomics research of legumes.

Chinese Bauhinia belongs to the subfamily Bauhiniaceae, which first diverged from Leguminosae, and is important for systematic genomic studies and new gene prediction in Leguminosae. To facilitate the genomic study of Leguminosae, the authors sequenced the genome of Chinese Bauhinia and obtained a genome of 352.84 Mb, which was assembled into seven pseudo-chromosomes and predicted 30,612 protein-coding genes (Fig. 1).

In contrast to the genomes of other legumes, there were no lineage-specific polyploid events in Bauhinia chinensis. Systematic genome analysis of 22 legumes and 11 angiosperms showed that many gene families were genealogically specific before and after legume diversification (Fig. 2).

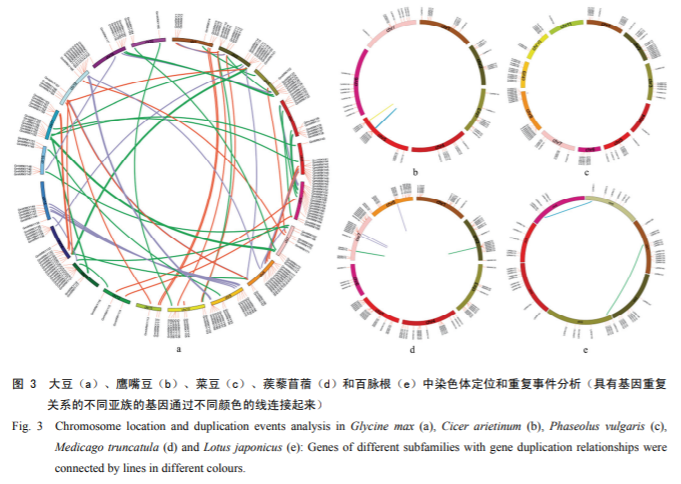
Further comparisons of multiple genomes of legumes revealed the presence of many de novo origin genes in different lineages of legumes, especially those that have undergone polyploidization (Fig. 3).

In this study, we sequenced the whole genome of Bauhinia sinensis, an important medicinal and horticultural plant widely distributed in southeastern China. Gene duplication events in the Chinese Bauhinia genome were found in different lineages of the legume family. In addition, hundreds of gained and lost gene families were identified in the evolutionary process of legumes, facilitating the inference of new genes, which contributes to the improvement of legume biodiversity. In addition, these well-annotated genes provide a reliable reference for further phylogenetic and genomic studies.