Astragalus sinicus (2n=2x=16), native to China and widely grown in South Asian countries such as Korea and Japan, belongs to the leguminous family of crops of the yellow sage family. Astragalus sinicus is an important green manure crop. Crop rotation during the winter fallow period can improve soil fertility by replacing 40% of mineral nitrogen fertilizer, thus effectively alleviating the problem of acidification of farmland soil by excessive inorganic nitrogen fertilizer. In addition, purslane is a source of high-quality pasture, bees, and an important herbal medicine. However, due to the lack of genomic information, the genetics and symbiotic nitrogen fixation studies of Zizyphus vulgaris lag far behind those of other leguminous crops.

Recently, Cao Weidong’s team from the Institute of Agricultural Resources and Agricultural Planning, Chinese Academy of Agricultural Sciences (CAAS) published an article “The chromosome-level genome assembly of Astragalus Sinicus and comparative genomic analyses provide new resources and insights for understanding legume-rhizobial interactions” in Plant Communications online. The chromosome-level genome assembly of Astragalus Sinicus and comparative genomic analyses provide new resources and insights for understanding legume-rhizobial interactions”. The study reported the first chromosome-level genome of Astragalus Sinicus, a leguminous green manure crop, which fills the gap of genomics research on leguminous green manure crops and provides new insights into the symbiosis between legumes and rhizobacteria.
The study used PacBio long-read-length data (144.11 Gb, ~230×), Illumina short-read-length data (74.58 Gb), and Hi-C data (64.62 Gb) to assemble the Zygomyrna genome, resulting in a final assembly of the Zygomyrna genome of 595.52 Mb, with a Contig N50 of 1.50 Mb, Scaffold N50 The Contig N50 was 1.50 Mb, the Scaffold N50 was 78.42 Mb, and the Hi-C mount rate was 96.66%. Transcriptome data from roots, stems, leaves, flowers and seeds were used to assist genome annotation, and a total of 34,253 protein-coding genes were annotated, with an average sequence length of 2.9 Kb.


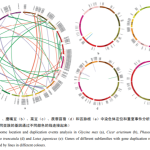
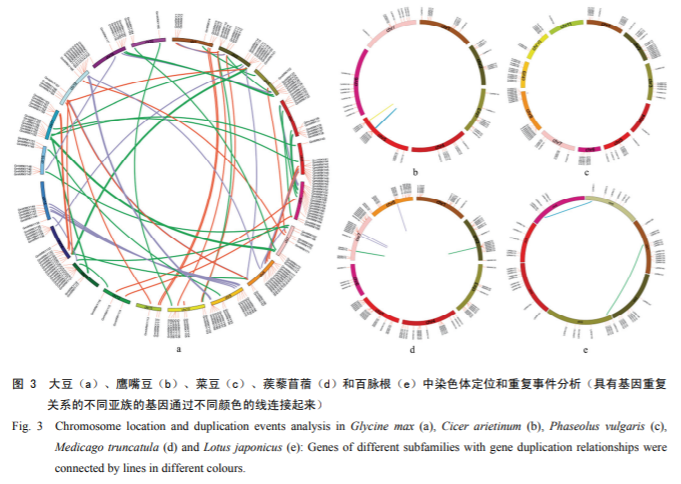
The researchers used a combination of ab initio and homology annotation methods to identify the repetitive sequences, and the Zi Yunying genome contained 59.84% of repetitive sequences, 96.97% of which were TEs. these LTRs underwent a dramatic amplification 500,000 years ago, which may have contributed to the expansion of the Zi Yunying genome. Meanwhile, the researchers also analyzed the genome evolution of 19 legumes and Arabidopsis thaliana and found that purslane diverged from licorice 26.4 million years ago and then from chickpea 19.1 million years ago. In addition, the researchers found that the whole-genome duplication events shared by Zygophyllum and Pteridophyllum subfamilies favored rhizome formation and hormonal regulation of symbiosis with rhizobia.

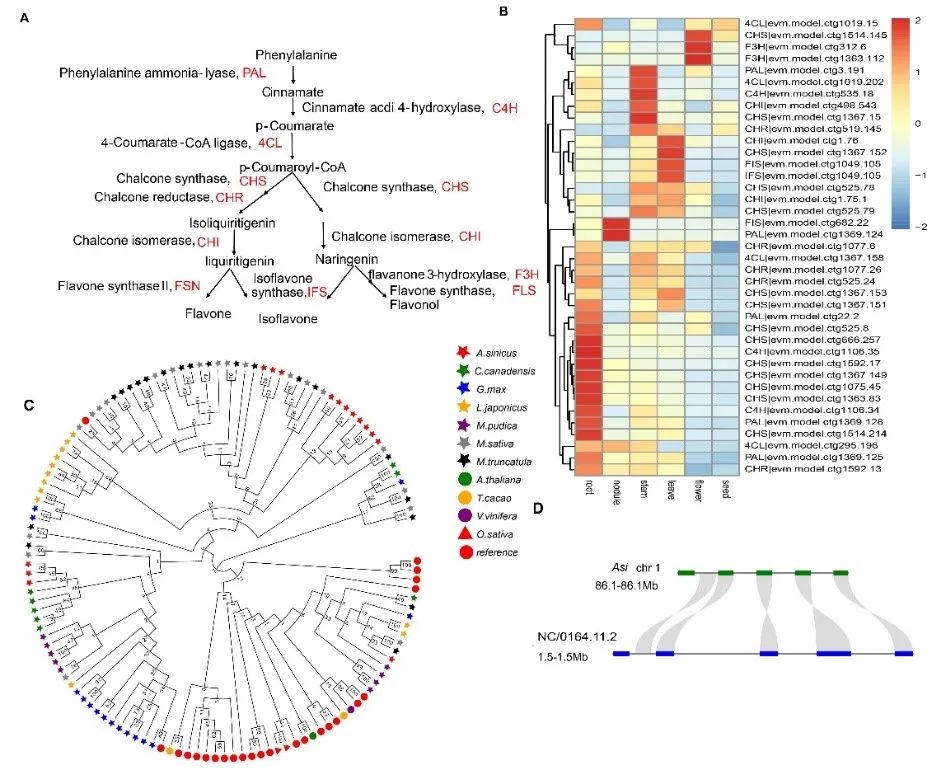
The results of gene family analysis showed that the chalcone synthase (CHS) gene family was the most significantly expanded gene family (15 copies) involved in the flavonoid biosynthesis pathway in Zygomyrrhiza glabra, and tandem duplication was responsible for the expansion of the CHS genes. Moreover, CHS genes have tissue expression specificity, and most CHS genes have higher expression in root tissues, which can provide high concentrations of flavonoids for rhizoma signaling.

Suppression of legume defense is a prerequisite for successful symbiotic development, and plant defense-associated R genes were enriched in roots and rhizomes, and R gene expression was higher in roots as a means of enhancing plant defense and counteracting rhizobacteria-induced immunosuppressio

The high-quality Zygomyrna genome provided by this institute will be an important resource for genetic and evolutionary studies, elucidating the genetic basis of Zygomyrna tuberculation and symbiosis, and providing theoretical support for accelerated genetic studies and molecular breeding in the future.