Mol Plant | Phylogenetic genomics reveals the origin of legume species divergence, genome polyploidy, and the hypothesis of nitrogen-fixing symbiotic evolution of rhizobia
The Leguminosae (Fabaceae) are one of the largest families of flowering plants, after Asteraceae and Orchidaceae. With its diverse forms and wide range of adaptations, it is one of the most successfully evolved groups of angiosperms. The Fabaceae comprise 765 genera and nearly 20,000 species worldwide, contributing 27% of global crop production. It contains a large number of species that have been cultivated and utilized globally and are economically important, such as common oilseed crops (soybean, peanut), vegetables or starches (kidney bean, pea, broad bean, cowpea, adzuki bean, black bean, lentil, mung bean, chickpea), important forage grasses (alfalfa, zoysia), valuable timber (sandalwood), commonly used traditional Chinese medicinal herbs (Astragalus, Glycyrrhiza glabra), and excellent ornamental trees for avenues of travel (Bauhinia, Bambusa, Mimosa, Bambusa, Mimosasaurus, Bambusia, Mimosa, Mimosa pudica). Bauhinia, Mimosa, Acacia, Phoenix canariensis, Rubia cordata, Acacia). Thus, the legume family provides a large number of important living resources for human beings. In addition, the symbiotic nitrogen fixation system of legumes and rhizobia is the biological nitrogen fixation system with the highest nitrogen fixation efficiency and the largest amount of nitrogen fixation in nature, and it is estimated that 17.2 × 107 nitrogen can be fixed annually. Effective utilization of symbiotic nitrogen fixation by legumes and rhizobia is of great significance to the sustainable development of agriculture. Therefore, exploring the evolutionary history of such a large taxon is not only of great scientific significance, but also of great socioeconomic value.
The Plant Evolutionary Biology Research Team at the School of Life Sciences, Fudan University has been conducting in-depth research on the broad framework of angiosperms, as well as representative families of economic, scientific, and ecological significance (e.g. Cruciferae, Asteraceae, Rosaceae, Cucurbitaceae, and Chrysanthemums) and ferns in the direction of plant systematics and evolution over the years. New Phytologist; Zeng et al. 2014, Nature Communications; Huang et al. 2016, Molecular Biology and Evolution; Xiang et al. 2017, Molecular Biology and Evolution; Zeng et al. 2017, New Phytologist; Ren et al. 2018, Molecular Plant; Qi et al. 2018, Molecular Phylogeny and Evolution; Guo et al. 2020, Molecular Plant; Zhang el al. 2020, Molecular Phylogeny and Evolution. Molecular Plant; Zhang el al. 2020, Molecular Biology and Evolution; Zhang et al, 2021, Journal of Integrative Plant Biology).
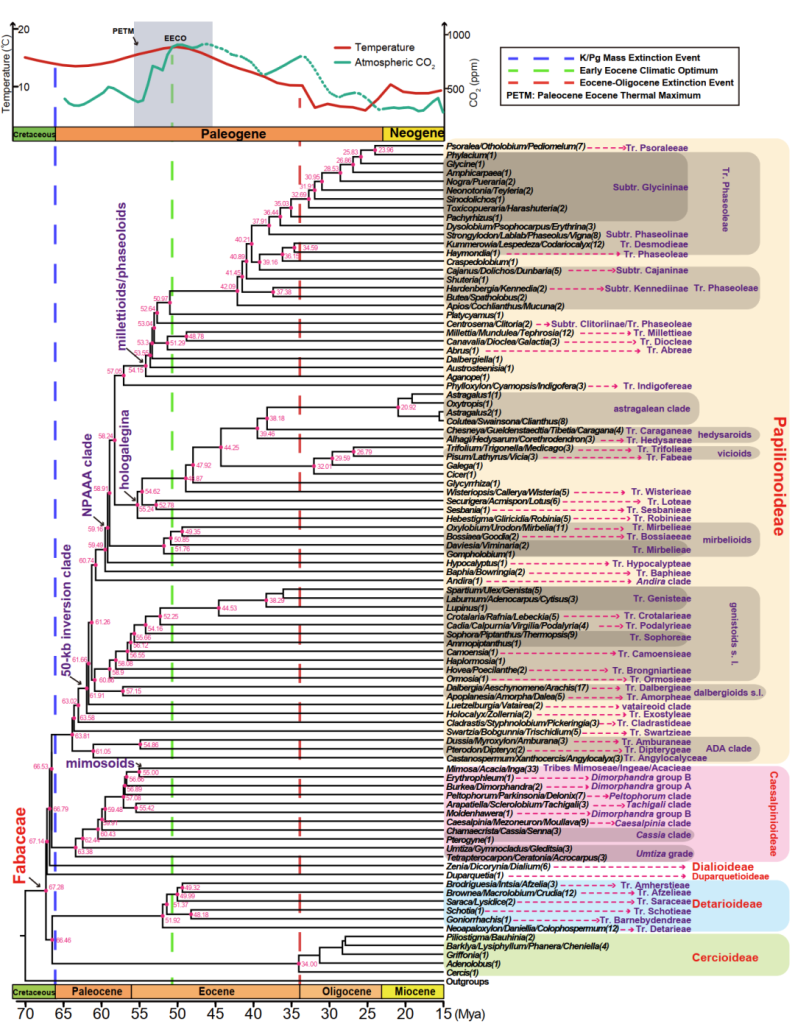
Figure 1, Correlation of legume origin and divergence times with changes in temperature and atmospheric carbon dioxide content over paleogeological time as analyzed by molecular clocks. The three vertical dotted lines in the figure indicate important geologic events, major clade or branch names are marked in violet, and the red numbers on the nodes in the evolutionary tree represent the divergence times of the crown groups.
In this legume group study, the team has obtained sequence data of more than 7 million nuclear gene transcripts from 391 species through extensive sampling, large-scale RNA sequencing or shallow DNA sequencing in international cooperation, which, together with other published gene sequence datasets, cover a total of 463 species in 59 clades or clades of all six subfamilies of the family Leguminosae. Based on phylogenetic analysis of eight low-copy orthologous nuclear gene sets, a highly supported legume phylogenetic tree was obtained, resolving some long-standing controversial or unresolved phylogenetic relationships. The results suggest that some clades or subclades need to be split and suggest the formation of new taxonomic units. For example, the three non-monophyletic clades (Mimoseae, Ingeae, and Acacieae) previously contained in the Mimosoids branch (mimosoids) provide evidence to consider reclassifying the above three clades into a single one under the designation of Mimoseae s.l. The new phylogenetic tree supports 100% of the existing phylogenetic relationships. The new phylogenetic tree supports 100% of the existing six subfamilies and resolves for the first time the deep phylogenetic relationships of the six subfamilies, with the Bauhinia subfamily (Cercidoideae) + Glycyrrhiza subfamily (Detarioideae) serving as the sister group to all the remaining legume families, followed by the differentiation of the monophyletic subfamily, the Ginger Bean subfamily (Duparquetioideae). Finally the acid olive bean subfamily (Dialioideae) as the sister group including the two largest subfamilies Caesalpinioideae and Papilionoideae branches.

Figure 2, a summary diagram showing a tree using flowers, fruits, and leaves of selected legume species to represent the phylogenetic relationships of the family Leguminosae.
The tree has six branches representing the six subfamilies of Leguminosae: the lower left to upper right of the tree shows the subfamily Detarioideae (Brownea grandiceps, Saraca dives), the subfamily Cercidoideae (Bauhinia chinensis, Bauhinia blakeana), the subfamily Cercidoideae (Bauhinia bracteatae), the subfamily Biloba (Bauhinia bracteatae), and the subfamily Biloba (Bauhinia bracteatae). Blakeana), Duparquetioideae (Duparquetia orchidacea), Dialioideae (Zenia insignis), Caesalpinioideae (Chamaecrista fasciculata, Mimosa pudica), and Cercidoideae (Brownea grandiceps, Saraca dives), Bauhiniaceae (Cercis chinensis, Bauhinia), and Caesalpinioideae (Chamaecrista fasciculata, Mimosa). The subfamily Dialioideae (Zenia insignis), the subfamily Caesalpinioideae (Chamaecrista fasciculata, Mimosa pudica, Acacia julibrissin, Calliandra haematocephala), and the subfamily Papilionoideae (Peanut Arachis hypogaea, Lupinus polyphyllus, Lupinus violetus, Lupinus viviparius). polyphyllus, alfalfa Medicago sativa, pea Pisum sativum, faba bean Vicia faba, soybean Glycine max, lentil Lablab purpureus, kidney bean Phaseolus vulgaris and cowpea Vigna unguiculata). The upper left infusion bag employs a metaphorical approach to show genome-wide duplication events in the major branches of the Leguminosae family, which, in descending order, are the most recent common ancestors of the Leguminosae ancestor, the Glycine max subfamily, the Bauhinia subfamily, the Acidolobaceae subfamily, the Cloudnut subfamily, and the Pteridium subfamily, all of which probably underwent a polyploidization event. Roots of an entire tree demonstrate rhizomes mediated by Rhizobium in the Leguminosae family. The spatial and temporal context of the figure shows the hypothesized asteroid impact on Earth 65 million years ago, which produced a globally-covered dust cloud that obscured the sky and drastically reduced global temperatures, leading to the fifth mass extinction event of species, including the dinosaurs. Flowers, pods and seeds of the legume family are scattered on the ground, and some of the beneficial insects and animals that may have facilitated legume pollination or seed dispersal are also shown, including ants, moths, butterflies, bees and squirrels.
A calibrated molecular clock based on 23 reliable ancient fossils reveals that the ancestor of the legume family originated about 67 million years ago, slightly before the Cretaceous-Paleogene boundary (K-Pg), the period of the “Great Dinosaur Extinction,” the fifth and most recent mass extinction event in Earth’s history. This period was the “Great Dinosaur Extinction”, the fifth and most recent mass extinction event in Earth’s history. Geological studies have shown that the main cause of this extinction event was probably a volcanic eruption caused by an asteroid impacting the Earth, a devastating event that caused a huge catastrophe for the Earth’s ecosystems, producing a dust cloud that covered the globe, obscuring the sky, while global temperatures dropped dramatically, and the cold ground temperatures made it difficult for seeds to sprout, which led to the extinction of about three fifths of all species during this period. extinction, including a wide range of nudibranchs and ferns, among others. So how did the legume family survive this catastrophe and flourish as one of the most successful groups of angiosperms?
This study identified 28 whole genome diploidization or triploidization events (WGDs/WGTs) in Leguminosae, including duplications that occurred in the ancestor of Leguminosae as well as in the ancestors of each of the five subfamilies. Furthermore, in response to the hypothesis of multiple heterologous polyploidizations among legume subfamilies proposed by Koenen et al. (Syst. Biol., 2020) based on data from eight legume species, a comparative analysis of data from 65 legume species including multiple genome sequences was performed, and results were obtained in support of polyploidization in the legume ancestor. Genome-wide duplications provided additional gene copies for processes such as responses to external biological stimuli, transmembrane transporter protein activity, and plasma membrane and protein metabolic processes, likely creating new drivers for adaptive evolution in legume species.
The results of the genome-wide duplication study implied that gene duplications in the legume ancestor likely provided the genetic molecular basis for symbiotic nitrogen fixation by rhizobia in the legume family. The team further analyzed the molecular evolutionary patterns of 30 families including genes that play key roles in important aspects of nitrogen-fixing rhizomes from 28 genomic data, and proposed a new hypothesis for the evolution of nitrogen fixation in legume rhizomes: namely, that a primary or secondary transfer from actinomycete-mediated to rhizobium-mediated symbionts (Microsymbiont Switch) occurred early in the legume. Internal deletions of two genes and duplications of multiple genes in tuberous legumes support the likelihood of a single symbiont transfer event in the ancestor of the legume family, however, the hypothesis of an earlier possible switch cannot be ruled out, e.g., in the common ancestor of the legume family. Meanwhile, gene family analyses showing the absence of different genes in non-tumor-bearing legumes support a parallel loss of nitrogen fixation in non-tumor-bearing legumes.
In short, this study suggests that the early genome-wide replication event in Leguminosae may have provided a rich genetic material base for stable and efficient nitrogen-fixing rhizomes, which provided intrinsic conditions for the derivation of a series of physiological and ecological traits. Meanwhile, the favorable climatic conditions in the middle Eocene as an external factor, the K-Pg species mass extinction provided more ecological niches for legume species and the stable and efficient nitrogen-fixing capacity of legumes synergistically contributed to the diversification process of legume species, which eventually evolved into a successful angiosperm group. This study provides an important perspective for our understanding of Cenozoic biodiversity (Cenozoic species diversity).
Yiyong Zhao, a PhD student at the School of Life Sciences, Fudan University, is the first author of the article, and Associate Researcher Jianxun Huang of Fudan University, Professor Hong Ma of Pennsylvania State University, and Researcher Tingshuang Yi of the Kunming Institute of Botany are the co-corresponding authors of the article. This research was co-funded by the National Natural Science Foundation of China, the State Key Laboratory of Genetic Engineering of Fudan University, the China Scholarship Council and the Pilot Program of the Chinese Academy of Sciences.
From WeChat https://mp.weixin.qq.com/s/2_c_AcGJ0UHkuWJzsyEMXQ
Share this content:


Post Comment